ABACUS strategy for protein NMR structure determination
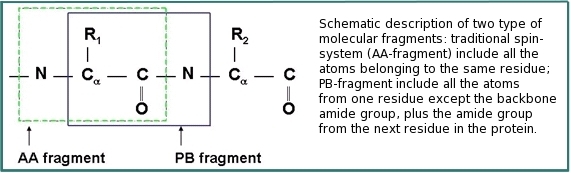
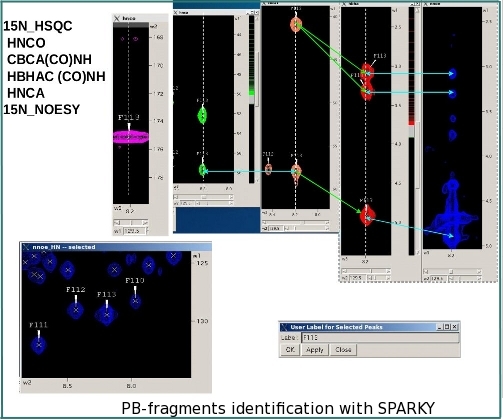
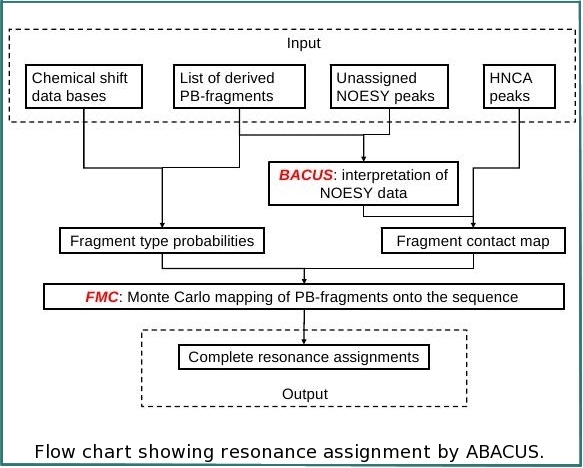
ABACUS (Applied BACUS) is a novel approach for protein structure determination that has been applied successfully for more than 20 NESG targets. ABACUS is characterized by use of BACUS, a procedure for automated probabilistic interpretation of NOESY spectra in terms of unassigned proton chemical shifts based on the known information on "connectivity" between proton resonances. BACUS is used in both the resonance assignment and structure calculation steps. The ABCUS is distinguished from conventional approaches to NMR structure determination mostly by its resonance assignment strategy. The resonance assignment procedure starts from grouping resonances in spin systems (PB-, or peptide bond, fragments) comprising correlated resonances from the side chain of residue i and the NH resonances of residue i+1 (see figures). HN-rooted PB spin-systems are collected from high sensitivity NMR correlation experiments (such as HNCO, CBCA(CO)NH, and HBHA(CO)NH) that transfer magnetization via the intervening peptide bond. Completion of PB-fragments with side-chain aliphatic resonances as well as identification of additional spin-systems (lacking HN resonances) is performed using HCCH-TOCSY and 13C-NOESY spectra. Sequence-specific assignment of PB-fragments is achieved using a Fragment Monte Carlo (FMC) stochastic search procedure. The scoring function used in the FMC procedure is based on both fragment amino acid typing (matching the spin system to amino acid types) and fragment contact map (reflecting which residue is next to which) derived from HNCA data, and the analysis of NOEs interpreted by BACUS.
Some features /advantages of the ABACUS protocol:
** It does not rely on sequential connectivities from less sensitive experiments such as HNCACB which most traditional sequential assignment procedures rely on
** Inter-residue sequential connectivities are established mainly from NOE data, which saves time at later stage in “trouble shooting” NOE and resonance assignments.
** Probabilistic nature of the ABACUS procedure provides measure of reliability of assignments, and therefore, the one can obtain a partial, yet highly reliable assignment (even when NMR data are sub-optimal) and knows where to focus manual intervention.
** It can make use of partial PB spin-systems (including those that lack an NH)
** It can efficiently identify manual errors in the input peak list.
How to get ABACUS
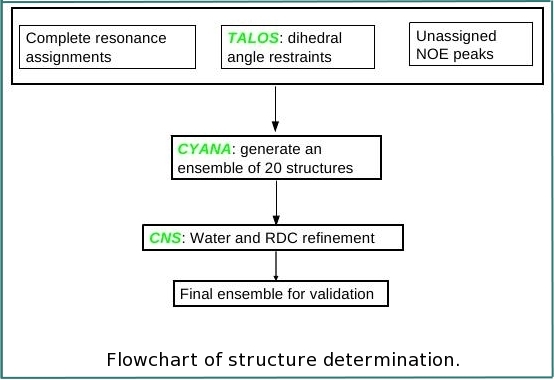
FMCGUI is a package of scripts with graphical interface that assists user to perform semi-automated resonance assignment using ABACUS approach as well as to set up structure calculations using CYANA and structure refinement using CNS.
FMCGUI is written in Python and Fortran for Linux OS. It requires Python, Tkinter, and Fortran packages to be installed on your system. In addition to this, FMCGUI make use of the following programs:
1. TALOS
2. PALES
3. RCI
4. CNS (version 1.3 is recommended)
Note that these programs are not required for resonance assignment using ABACUS approach. They are used only for structure calculation/refinement.
FMCGUI distribution for Linux can be downloaded HERE.
To install FMCGUI, unpack the downloaded file and follow the installation instructions in the README.
The description of FMCGUI commands and how to use it could be found on the NESG NMR-Wiki site.
Citation Information
Lemak A., Gutmanas A., Chitayat S., Karra M., Farès C., Sunnerhagen M., and Arrowsmith CH. "A novel strategy for resonance assignment and protein structure determination". Journal of biomolecular NMR 49:27-38, 2011.
Lemak, A., Steren, C.A., Arrowsmith, C.H., and Llinas M. "Sequence specific resonance assignment via Multicanonical Monte Carlo search using an ABACUS approach" Journal of biomolecular NMR 41:29-41, 2008.
Grishaev, A., Steren, C.A., Wu, B., Pineda-Lucena, A., Arrowsmith, C. and Llinas, M. "ABACUS, a direct method for protein NMR structure computation via assembly of fragments" Proteins 61: 36-43, 2005.
